A guided tour through LORETA – Source localization in BrainVision Analyzer 2!
by Dr.-Ing. Kidist Mideksa
Scientific Consultant (Brain Products)
![]() What do I need in order to include source analysis as part of my analysis pipeline? This article provides you with the solution mainly focusing on LORETA. We will also walk you through the requirements prior to applying LORETA so that you will be able to make the best usage of LORETA in BrainVision Analyzer 2.
What do I need in order to include source analysis as part of my analysis pipeline? This article provides you with the solution mainly focusing on LORETA. We will also walk you through the requirements prior to applying LORETA so that you will be able to make the best usage of LORETA in BrainVision Analyzer 2.
Overview
Introduction
Why source analysis? EEG recordings at the surface of the scalp represent the summed electrical activity coming from the different parts of the brain. This electrical activity is generated by large numbers of neurons with electrical charges moving within the central nervous system and causing small electrical currents. The amount of current flow is characterized by the volume conduction, i.e., the electromagnetic properties of the tissues inside the brain.
There have been many algorithms developed so far for processing these signals at the scalp-level. The operations include, but are not limited to, time-domain analysis, frequency-domain analysis, and time-frequency analysis. Signals measured on the scalp, however, do not necessarily represent the true neuronal activity in the brain. That is, due to the volume conduction effect, electric currents spread over a relatively large distance from the sources to the electrodes, making EEG measurements possible in the first place. However, the underlying neural dynamics cannot be revealed because of considerable attenuation, mixing and smearing of the neuroelectric fields arising from different sources while spreading through the tissues of the brain until they reach at the scalp. Thus, there is a need for analysis techniques that enable backward tracing of the active brain regions. Using source analysis you will be able to estimate the location and dynamics of the underlying neuronal generators responsible for the electrical potentials measured on the scalp.
Source analysis consists of solving the so-called forward problem and inverse problem. Given the physiological sources, one can calculate the generated electrical potentials using classical electrodynamics equations. This is known as solving the forward problem. When proceeding in the opposite direction one can infer where the underlying physiological sources are located by estimating the current distribution from the measured electrical potential. This is known as solving the inverse problem. Different forward and inverse methods offer an opportunity to select the most appropriate algorithm for a given experiment or research paradigm. In this article we will mainly focus on LORETA (Low Resolution Brain Electromagnetic Tomography), which is one of the most established and widely used current density reconstruction algorithms (Pascual-Marqui et al., 2002). It enables you to estimate distributed activity throughout the brain volume by decomposing the overlapping EEG voltage patterns into their underlying sources and localizing them within the brain. LORETA is not only answering the question of where the sources are located, but it also provides you with the time courses of each source.
Prerequisites for LORETA
As in any other inverse method, source estimation with the help of LORETA also requires several sophisticated steps, grouped into the forward and inverse problem. No worries, all these steps are implicitly taken care of in the implementation of LORETA in Analyzer 2. Thus, what is expected from you are mainly two things:
- Valid head coordinates. If your data does not contain valid head coordinates (standard or realistic), you can adopt the default coordinates as defined internally in Analyzer 2, as long as your channel names match the international 10-20 electrode system and its derivatives (i.e., 10-10 and 10-5 systems). Moreover, it is also recommended to have a uniform distribution of the electrodes throughout the scalp with the number of electrodes being at least 64 or higher (Michel CM et al., 2004). The coordinate system used by Analyzer 2 is illustrated in the User Manual (Appendix C Electrode coordinate system).
- Voltage measurements (EEG data). The most important pre-requisite which is highly specific to Analyzer’s LORETA is that only time-domain voltage values are valid input for the algorithm. Moreover, as in any kind of scalp-level analyses, for e.g., ERP analysis, source analysis also benefits from a high signal-to-noise ratio, which can be achieved by having well pre-processed data.
This is the only prior information that is required from your side. However, if you are curious to know more about the implicit mechanisms of the forward and inverse problem, we will provide you with the basic ingredients without going too deeply into the mathematics behind it. To start with, consider that you have two domains: one being the electrode space where you have the voltage measurements and the second one being the source space, where the neuronal current sources are confined inside the brain. As mentioned above, the forward problem computes the electrical potentials based on the information about channel coordinates from the electrode space and the source parameters (position, orientation and magnitude of the neuronal current sources) from the source space. This computation entails modeling of 1) the neuronal current sources and its spatial distribution within the brain as well as 2) the volume conductor medium.
- Source model: a pre-defined source space is important to model sources. In Analyzer 2, the source space is restricted to the regions of the cortical gray matter and hippocampus in the Talairach atlas, discretized into a total of 2394 voxels at 7 mm spatial resolution. These sources are then modeled using equivalent current dipoles.
- Volume conductor model (anatomical and head models): it describes the geometrical and conductive properties of the head. In Analyzer 2, we offer a standard brain which is based on the MNI-305 brain template. The MNI images are co-registered to the Talairach brain atlas to map the detailed brain structures into the MNI space. Moreover, our LORETA implementation is based on a 3-shell spherical head model. This head model is fitted to the MNI brain template, which has already been co-registered to the Talairach brain atlas.
In addition, the electrodes shall be projected to the co-registered spherical head model. Once all these ingredients are brought to the same coordinate system, the lead-field matrix, [Nelectrodes x 3Msources] (3 represents the X, Y and Z components of the sources), is computed. This matrix serves as a mapping mechanism from the neuronal current sources within the brain to the electrical potentials on the head surface.
All of the above steps are necessary pre-requisites for solving the inverse problem. The inverse problem aims to minimize the difference between the true voltage measurements and the calculated electrical potentials, as provided by the forward solution. Strikingly, there is an infinite number of source spatial configurations giving rise to the same voltage distribution. Therefore, the inverse problem in general does not have a unique solution, unless certain assumptions and mathematical constraints are imposed. In the case of LORETA, a unique inverse solution is achieved by imposing a constraint on the spatial configurations of the neuronal current sources, i.e., LORETA aims to find the smoothest spatial distributon within the source space. (Pascual-Marqui et al., 1994 and Pascual-Marqui et al., 1999). Finally, the unique inverse matrix, [3Msources x Nelectrodes], fulfills all the conditions mentioned above, which relates the true voltage measurements to the estimated neuronal current sources.
Accessing LORETA
There are mainly two approaches to access LORETA in Analyzer 2. To decide which approach suits you best, let’s start asking the following questions:
- Are you aiming at performing an exploratory analysis to identify the main brain areas underlying a given ERP/EEG pattern? Are you aiming to infer the Regions of Interest (ROIs)? In this case, you are looking for the LORETA transient view. The transient LORETA is available via the right-click Transient Views menu.
- After inferring your desired ROIs in the source space with the transient LORETA, are you now aiming at extracting the underlying neural dynamics as ROI-specific time series? Do you want to create ROIs based on other (e.g., fMRI) studies and/or load ROIs from the pre-defined source space (the MNI voxels)? Then you are looking for the LORETA transform which is available via the Transformations ribbon (Transformations > Special Signal Processing > LORETA).
Creating Regions of Interest (ROI)
In this section, we will show you how to create ROIs and the possible computations that can be performed on the created ROIs.
As mentioned above, the LORETA transform enables you to create and/or load ROIs. ROIs can be created by clicking on the Insert ROI button in the LORETA dialog, which will generate nodes automatically labeled as NewROI01, NewROI02,… depending on the number of times you insert a new ROI. You then have the possibility to edit the ROI name.
A single ROI node can be constructed in various different ways:
-
- Only one voxel associated to a specific brain region: A single voxel can be created with the option Add Nearest Voxel. Let’s say during the exploratory analysis using the transient LORETA, you found out that the strongest source activation was located at the MNI coordinate (X = -3, Y = -74, Z = 29) and now you want to explore the dynamics of this single voxel. To do that, you first insert a ROI node and then using your mouse, fine tune the position of the yellow sliders to the desired position in the MNI brain image. Then click on the option Add Nearest Voxel button as shown in Fig. 1. It will give you the best matching location of what you have defined using the yellow sliders.
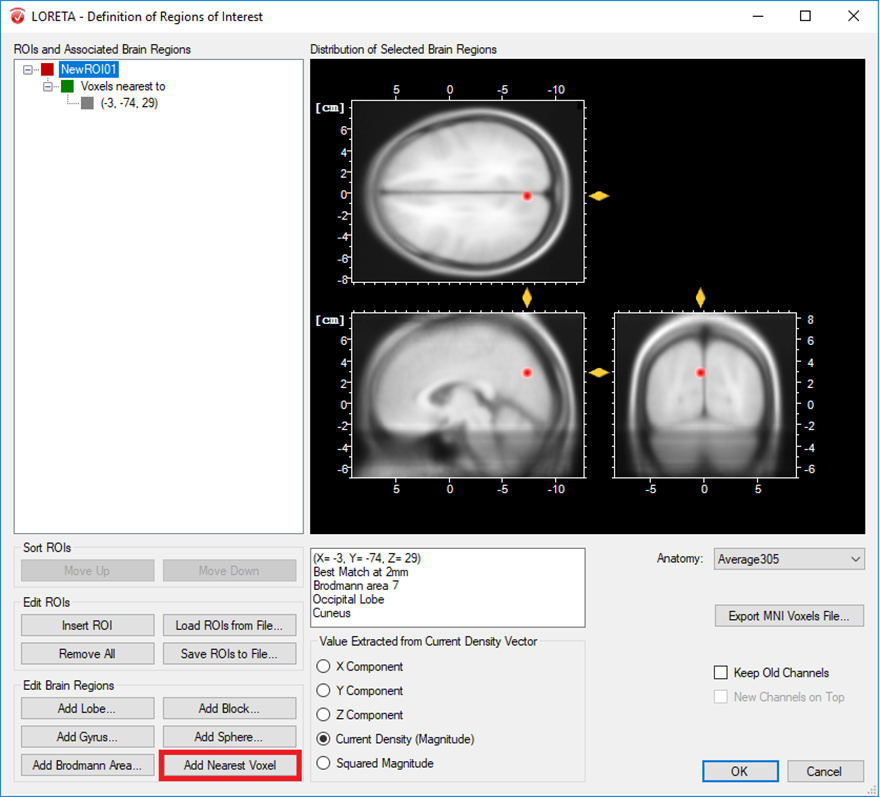
Fig. 1: Creating a single ROI associated to a single voxel from a pre-defined source space database.
The other possibility is that you already have a-priori information about the desired voxel from other studies and would like to create it. This is possible by making use of the pre-defined source space database. This database can be accessed from the LORETA dialog by clicking on the Export MNI Voxels File button. If you then open the saved *.csv file, you will see detailed information about all the pre-defined 2394 voxels. So, let’s say that the a-priori information you have is the MNI coordinate (X = -52, Y = 3, Z = -41). You can search for this coordinate in the file and once you find the desired voxel, change its corresponding ROI-number (last column) to 1 as shown in Fig. 2. Please also make sure to change the total number of ROIs (2nd line of the *.csv file) from 0 to the desired number of ROIs (for e.g., to 1 in this case). You can then save it and go back to the LORETA dialog to load the modified *.csv file using the button Load ROIs from File. Generally, you can create K number of ROIs associated to a single voxel by simply incrementing the ROI-numbering from 0 to 1,2,3,…K and finally changing the total number of ROIs (2nd line of the *.csv file) to K. Fig. 3 illustrates how to create 4 ROIs, each associated with a single voxel.
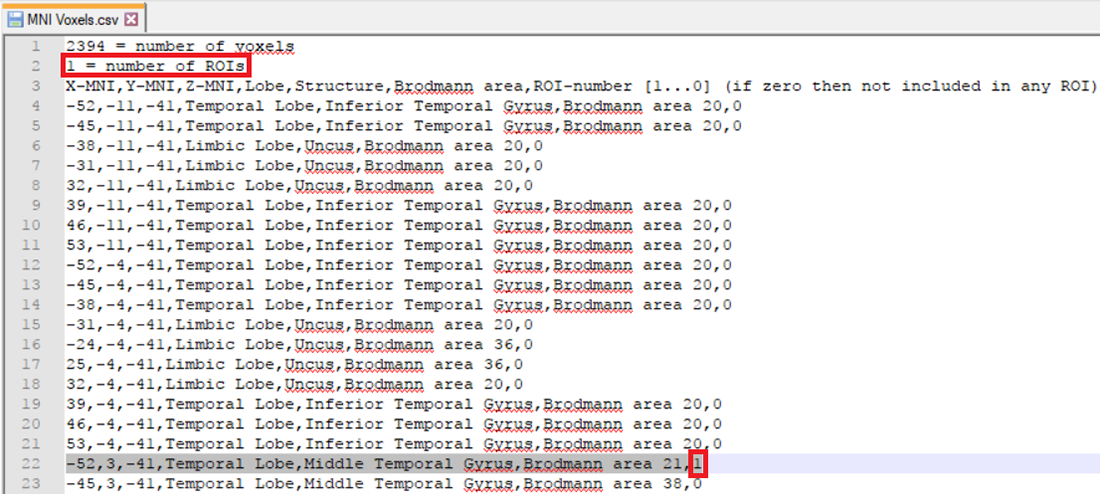
Fig. 2: Creating a single ROI associated to a single voxel from a pre-defined source space database.
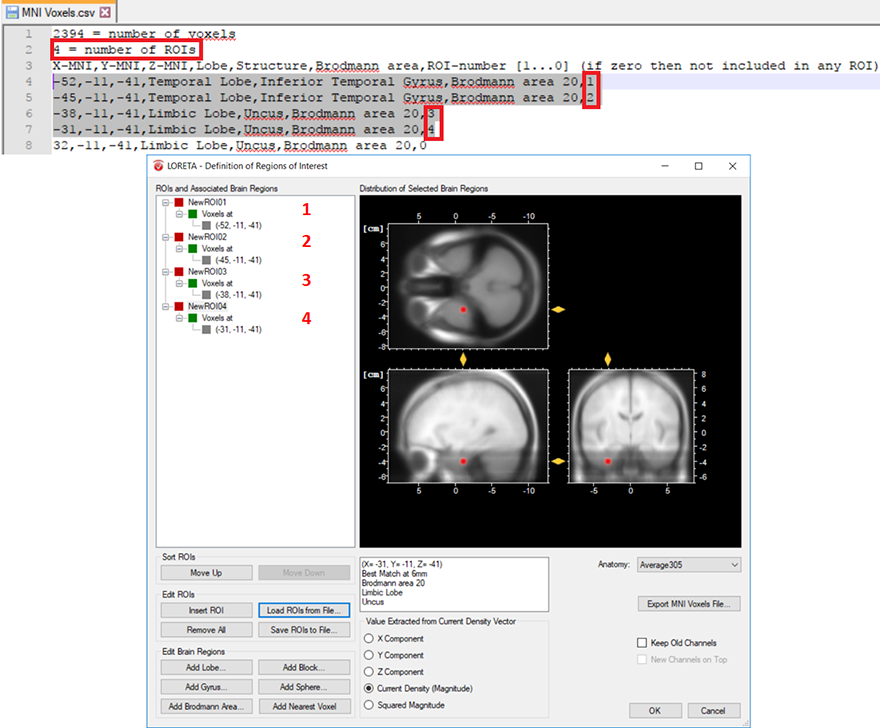
Fig. 3: Creating 4 ROIs, each associated with a single voxel from the pre-defined source space database. The voxel associated with the ROI labeled as NewROI04 can be visualized with the red area in the MNI brain image.
-
- Cluster of voxels associated with a specific brain region: In a similar way, you can also create several voxels associated with a single ROI. This is achieved by simply labeling all desired voxels with the same ROI-numbering scheme. Then change the total number of ROIs from 0 to 1, if a single ROI is desired as shown in Fig. 4. For instance, you can select L voxels and group them to the first ROI, another M voxels to the second ROI and so on. This can be achieved by labeling all the L voxels with the same ROI-numbering scheme (e.g., labeled as 1 to group them in the first ROI) and all the M voxels (e.g., labeled as 2 to group them in the second ROI) and so on. Finally, change the total number of ROIs to the desired number of ROIs you would like to create.
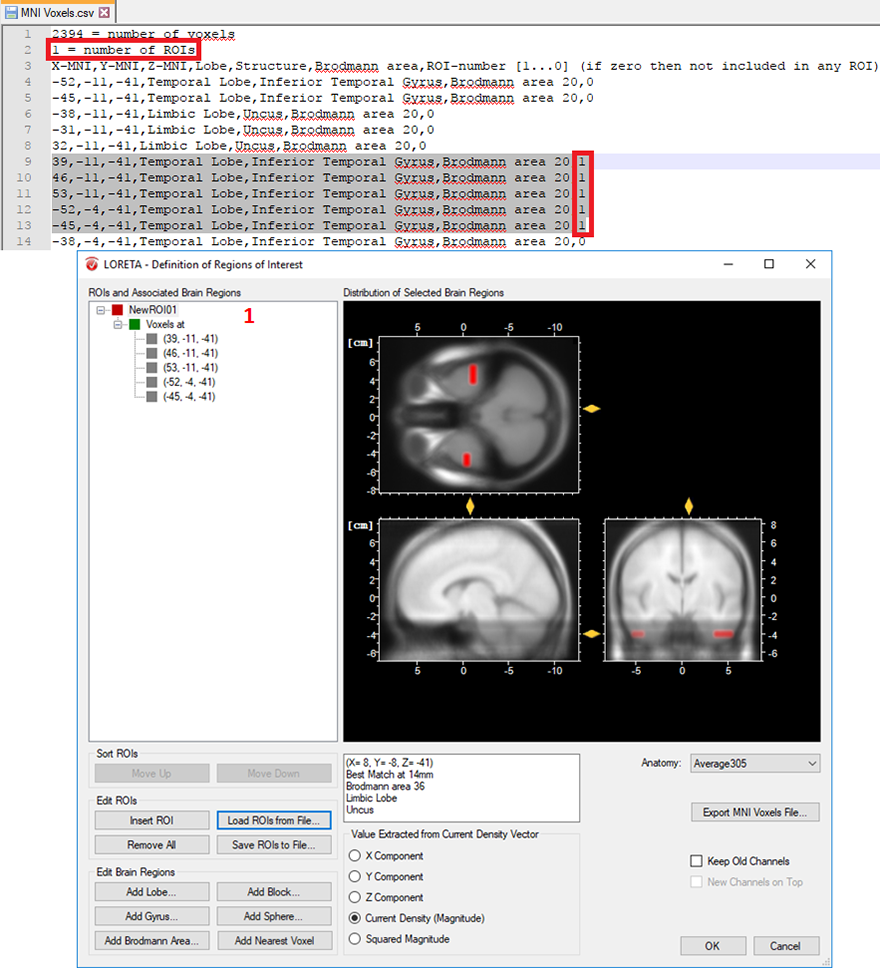
Fig. 4: Creating a single ROI associated to 5 voxels from the pre-defined source space database.
-
- Cluster of voxels associated to different regions of the brain: Different structures can also be associated to a single ROI either based on lobes, gyri or Brodmann areas by clicking on the Add Lobe, Add Gyrus, Add Brodmann Area button, respectively. CAUTION! Unless there is a rationale behind creating such a ROI, it is not recommended to combine different regions of the brain in a single ROI due to the computational reasons explained below in the Output Measures section.
Once you are done with defining your ROIs, you can make use of the Save ROIs to File option, which will enable you to save the created ROI tree to an *.xml file. This will allow you to conveniently share your ROI tree across different workspaces in Analyzer 2.
See more, do more: Make use of our 88-ROIs file!
So far, you may have noticed that creating and labeling ROIs, as well as associating brain areas to existing ROIs entails a considerable amount of manual work. How would you proceed if you are interested in an automatic creation of ROIs associated to a particular brain parcellation? You can imagine how challenging it is to manually define a ROI for each parcel either by editing the pre-defined source space database, or using the options in the group Edit Brain Regions of the LORETA transform. For your convenience, we have taken over this tedious job and created an *.xml file containing 88 ROIs/parcels associated to Brodmann areas, Amygdala and Hippocampus, separately for the left and right hemisphere. You can download the file below and with the help of the option Load ROIs from File you will be able to import all the 88 ROIs in one go. In the same way automatic ROI definition for other brain parcellations can be achieved. In case you need further assistance on how to do it, please feel free to contact us.
Output Measures
After the successful creation of ROIs, the next step will be to estimate the current source density within the specified ROI. The LORETA dialog provides you with several output options. You have the possibility to compute for each time point the mean of the (X, Y, Z) component of the current density vector separately, the Magnitude of the current source density, or its Squared Magnitude within the specified ROI.
Tip: Applying different output options for different ROIs
The default output is the Current Density (Magnitude). In case you want different output options to be applied for the different ROIs, please make sure to select the desired output option for each ROI, separately.
Finishing the LORETA dialog will create a new history node named LORETA in the History Tree®. It contains new LORETA channel(s), representing the temporal dynamics of the neuronal current source(s) corresponding to the created ROI(s).
Tip: What to do if the resulting source time-series looks like a flat line
Don’t worry if you see the resulting source time-series looking like a flat line. This is due to the fact that current source density values have indeed very small but non-zero values. In addition to that, the scale used in Analyzer 2 to display signals basically fits the voltage value of a typical EEG, and is not appropriate to display the source strength values. Thus, please make sure to increase the scaling of the amplitude either by using ctrl+up arrow keys of your keyboard or using the Scale Up Amplitude button from the toolbar.
Tip: Visualizing both – the EEG and the LORETA source signals
If you selected the check box Keep Old Channels in the LORETA dialog, this will add the LORETA channel(s) to the original set of EEG channels. This enables you to compare the EEG signal(s) to the corresponding LORETA source signal(s). To visualize both signals, you can specifically select and rescale only the LORETA channel(s) by clicking on the Scaling Range button from the toolbar and choose the option “Scale Selected”.
The current density values obtained from the transient LORETA may differ from the results of the LORETA transform. Estimation of a source with the transient LORETA is done for the selected time-interval. So, what you will obtain for each displayed voxel is the magnitude of the vectorial mean of all current density vectors associated with all time points included in the selected time-interval . As illustrated in Fig. 5, what you see in the status bar of the LORETA transient view is the maximum current density (magnitude) (0.0401 µA/mm2), i.e., the mean of all current density vectors within the selected time-interval (61 ms – 131 ms) associated to a specific voxel located at (X = 4, Y = -81, Z = 8).
Tip: Making sure that the maximum current density is displayed
To make sure that the maximum current density is displayed, right-click anywhere in the dockable window of the transient LORETA and make sure that the option Lock Navigation to Maximum is checked. By default, this option is selected. With this setting, the yellow sliders will always point to the position of the voxel with the maximal activation, and at the same time its current density value is displayed in the status bar.
Tip: Exploring the complete 3D current source density in a 2D by 2D manner
The option Show Horizontal Slices gives you the opportunity to explore the complete 3D current source density distribution in a 2D by 2D manner.
Tip: Adjusting the scale of the current source density values
You have the possibility of adjusting the scale of the current source density values via the Settings dialog box (accessible by clicking on the option Settings). The option Automatic Scaling helps you detect where the focal point of the maximum current source density value lies. Based on this information, you can then use the Manual Scaling option to clip the current source density values above a certain threshold (i.e., concentrated around the maximum current source density value). This will enable you to display the largest current source density value only.
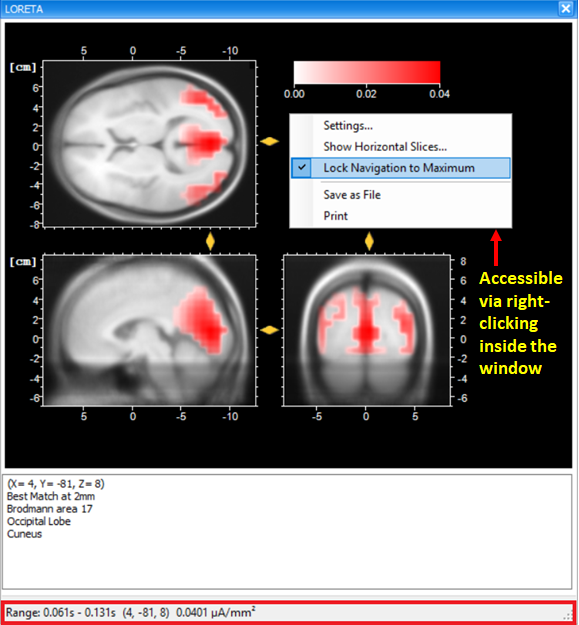
Fig. 5: Dockable window of the transient LORETA showing source activation map for the selected time-interval.
On the other hand, the LORETA transform provides you with the output measures for each time point. Analogous to the computation of the vectorial mean across time points based on the selection of the time-interval, the LORETA transform also computes the vectorial mean across multiple voxels, when multiple voxels are included within a single ROI. So, what you will obtain for a single LORETA channel is a source signal containing, for each time point, the average current density across all the voxels within a ROI, as shown in Fig. 6. This is the reason why we said to take care when adding different brain areas to a single ROI. Especially when creating ROIs comprising large brain areas, the current density vectors across all voxels may point in different opposing directions, and thus their contribution to the vectorial mean cancels out, which might not be desired. So, if extracting source signals from different regions of the brain is desired, we do recommend to create separate ROIs for each region and aggregate only the desired voxels to a ROI associated to a particular region, as illustrated in Fig. 7.
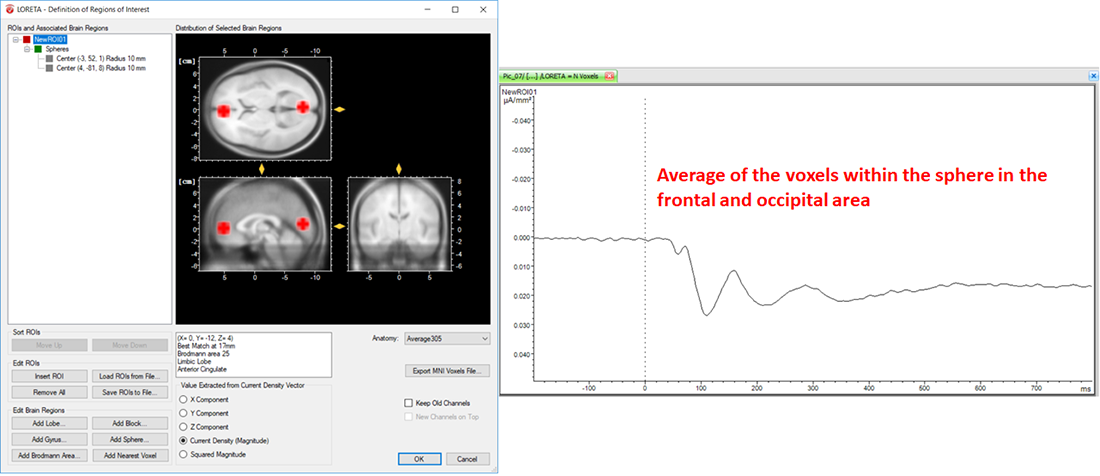
Fig. 6: Effect of including different structures of the brain within a single ROI along with its corresponding source signal depicted in one LORETA channel (NewROI01).
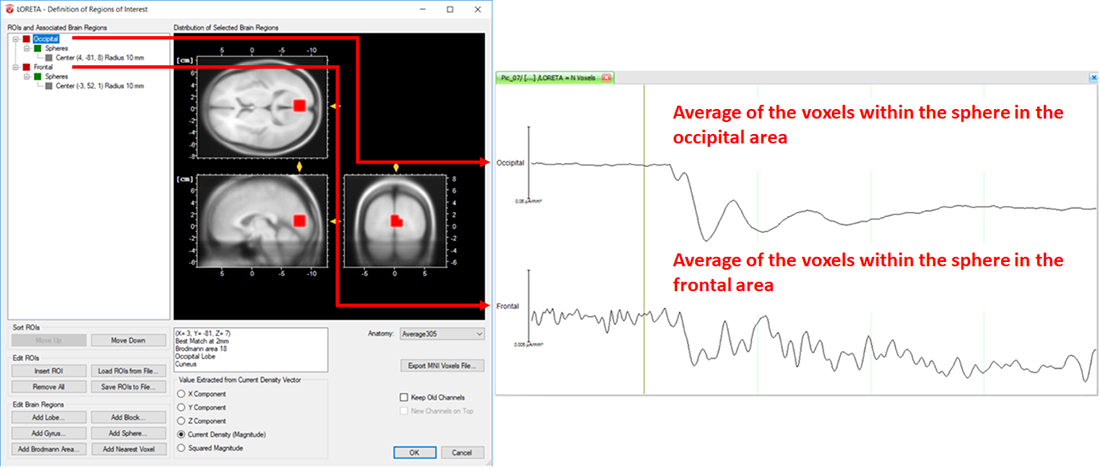
Fig. 7: Separate ROIs for the different regions of the brain along with its corresponding source signal depicted in separate LORETA channels (Occipital and Frontal).
Measure Extraction and Export
At this final stage you might be interested in exporting the results obtained from the LORETA transform for further statistical analyses. We will briefly provide you with some valuable tips about the measures that can be extracted from the source time series and the ways to extract them.
Exporting current source density values
Current source density values for each time point can be easily exported by applying the Generic Data Export module (Export > Node Export > Generic Data) on the LORETA node. The exported file will be stored under the “Export” folder of the current workspace.
Tip: Avoiding the loss of values by the rounding effect while exporting
Since current source density values are typically very small, in order to make sure that you don’t loose any value by the rounding effect while exporting, please make sure to increase the precision (e.g., 10 or more). The precision can be adjusted using the option “Use Custom Precision” in the Generic Data Export dialog.
Area measures
Are you looking for a way to export area information or the mean activity around the maximum source signal? The Area Information Export module (Export > Multiple Export > Area Information) enables you to compute and export area measures for the desired time interval.
Finding the peak current source density value
Are you interested in pinpointing the maximum and/or minimum current source density value of the source signal and its corresponding latency for each ROI? If so, you can make use of the Peak Detection transformation (Transformations > Segment Analysis Functions > Result Evaluation > Peak Detection). It enables you to detect and mark the maximum and/or minimum current source density value within the desired time interval. After detection, you might want to export the corresponding current source density value and latency for each ROI. The Peak Information Export module (Export > Multiple Export > Peak Information) enables you to export the detected maximum and/or minimum current source density value(s) and time point(s), separately for each ROI.
Summary
It is our pleasure to invite you to make use of the two modes of LORETA, i.e., the LORETA transient view and LORETA transform in Analyzer 2. As you can see they complement each other. With the help of the transient LORETA you can inspect your EEG data and make a first impression about the location of the activated area. Once you have an impression about the location, you can infer your ROIs and use the LORETA transform to create the ROIs and estimate the current source density, as well as extract the source time series. We hope that you will broaden your research questions beyond scalp domain analyses by diving into the source domain analysis. Further detailed information about the two modes of LORETA as well as the different Export modules can be found in the BrainVision Analyzer 2 User Manual. For a step by step protocol on how to use LORETA, we would like to invite you to have a look at the pdf slides of our webinar on “Introduction to Source Analysis – Distributed Source Imaging Using LORETA”. If you still can’t find what you are looking for, drop us a line at support@brainproducts.com. We will be happy to help!
References
[1] Pascual-Marqui RD, Michel CM, Lehmann D (1994).
Low resolution electromagnetic tomography: a new method for localizing electrical activity in the brain.
International Journal of Psychophysiology, 18:49-65.[2] Pascual-Marqui RD, Lehmann D, Koenig T, Kochi K, Merlo MCG, et al. (1999).
Low resolution brain electromagnetic tomography (LORETA) functional imaging in acute, neuroleptic-naive, first-episode, productive schizophrenia.
Psychiatry Res., 90: 169–179.[3] Pascual-Marqui RD, Esslen M, Kochi K, Lehmann D (2002).
Functional imaging with low resolution brain electromagnetic tomography (LORETA): a review.
Methods & Findings in Experimental & Clinical Pharmacology, 24C:91-95.[4] Michel CM, Murray MM, Lantz G, Gonzalez S, Spinelli L, et al. (2004).
EEG source imaging.
Clinical Neurophysiology, 115(10), 2195-2222.

